Web Application for Protein Structure Prediction
Posted on March 6, 2025 (Last modified on March 9, 2025) • 4 min read • 780 wordsAlphaFold prediction tools in a browser.

Introduction
Protein structure prediction is a fundamental aspect of bioinformatics and molecular biology, with significant applications in drug discovery, disease research, and synthetic biology. However, traditional computational methods can be complex and resource-intensive, making them difficult for many researchers to access.
To overcome these challenges, advanced computational engines have been developed to predict protein structures efficiently using computer algorithms. One of the most successful engines is AlphaFold, created by developers at Google DeepMind.
Our platform incorporates AlphaFold models, including AlphaFold2 and AlphaFold3, along with optimized versions of ColabFold, OmegaFold, and ESMFold. These tools usually operate through terminal scripts, which can be difficult for researchers without a computer science background. To address this, our platform offers a user-friendly graphical interface that makes it easier for users to execute predictions.
Why Protein Structure Prediction Matters
Proteins are the fundamental building blocks of all living organisms, and their functions are closely linked to their three-dimensional structures. Gaining insights into protein structures is crucial for drug discovery, as it allows researchers to identify potential drug targets and design new medications. Additionally, in disease research, understanding these structures aids in determining how mutations can impact protein function.
Traditional experimental methods, such as X-ray crystallography and cryo-electron microscopy, are time-consuming and expensive. Computational tools help fill this gap by providing faster and more accessible predictions.
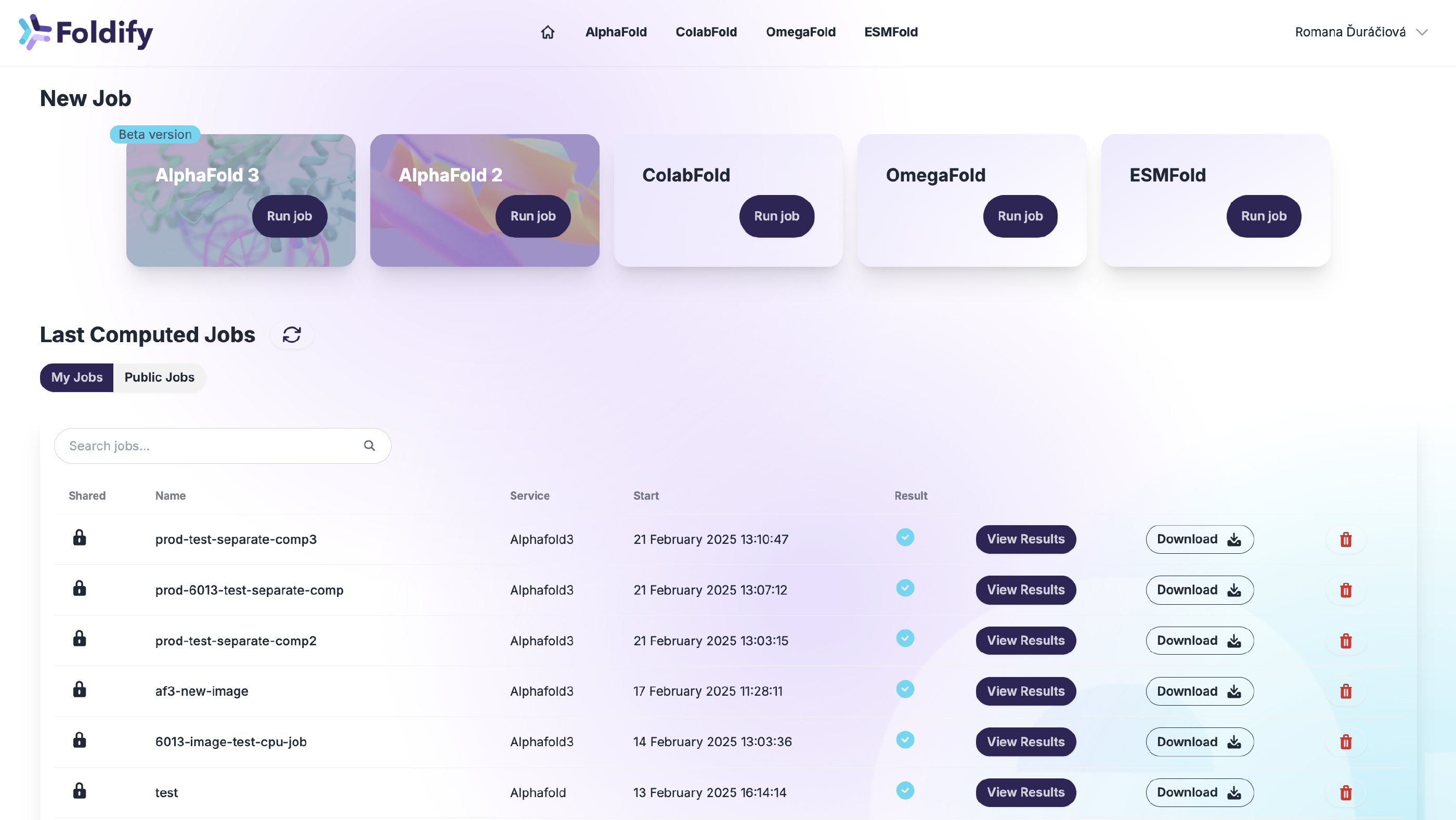
Overview of the web application
Foldify, our web application, streamlines the protein structure prediction process, making it faster and more user-friendly. It also provides real-time feedback to guide users in case of incorrect input. Key features of the application include:
Multiple Prediction Models in One Place
The Foldify platform offers a variety of folding tools, including AlphaFold3, AlphaFold2, ColabFold, and others. This provides users with the flexibility to select the tools that best fit their specific needs, unlike the original AlphaFold Server from Google, which only supports AlphaFold3.
User-Friendly Interface
Foldify provides a user-friendly graphic interface for form submissions, eliminating the need for complicated command-line inputs. The required data is minimal, and each input format is clearly explained with guiding instructions and helpful hints. Additionally, the system offers error feedback and input validation before you submit your data for computation to prevent the most common mistakes.
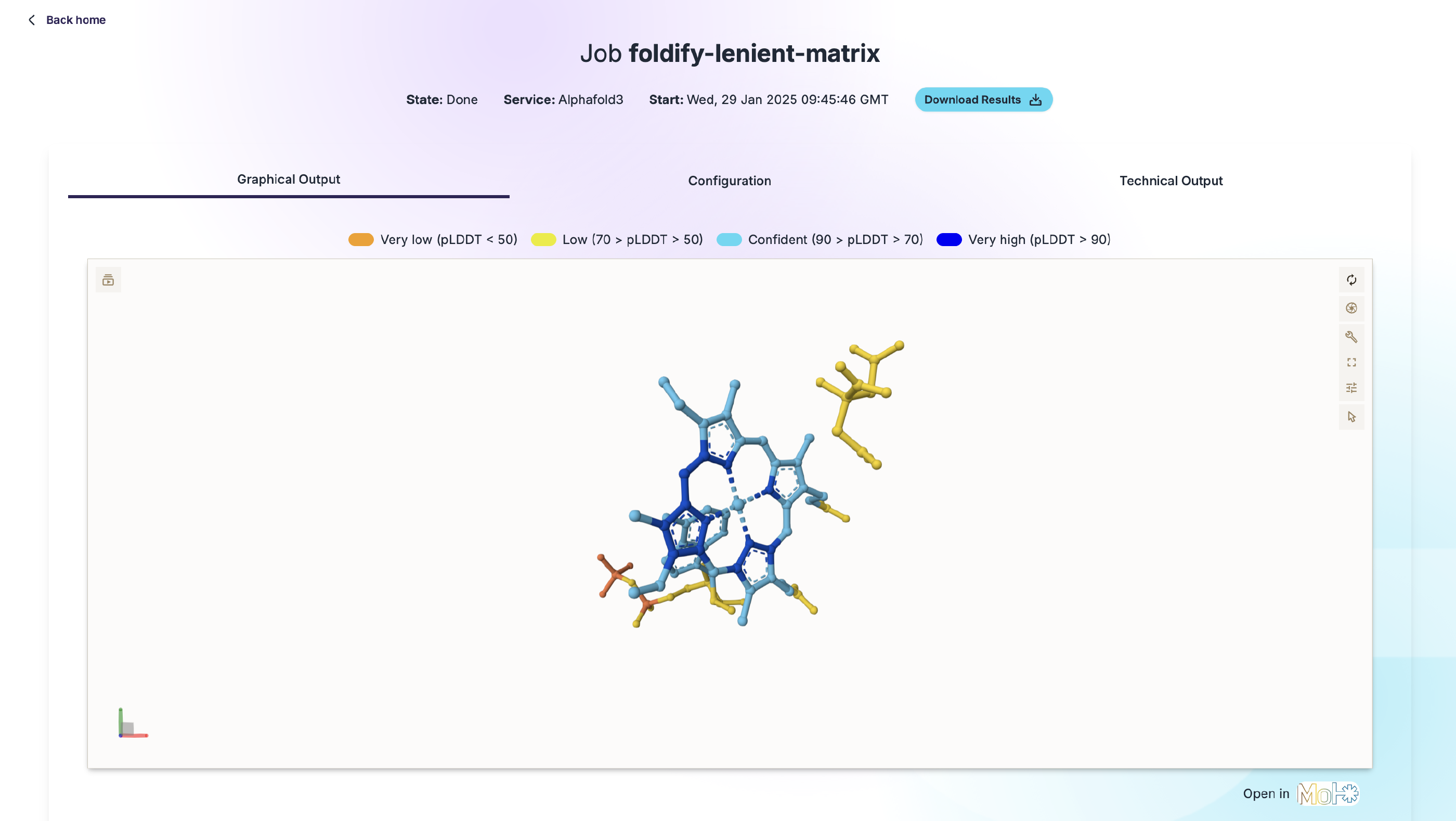
Built-In 3D Visualization
View and analyze predicted protein structures directly in your browser with the built-in Mol* Viewer. There’s no need to download large files for visualizations. With Foldify, you can easily view protein structures online using the popular Mol* visualization toolkit, which is also utilized by UniProt, the world’s leading resource for protein sequences.
Real-Time Monitoring
Monitor prediction progress in real time through a status indicator on your Dashboard. Users can check job status, view updates in console logs within the detailed computation view, and receive email notifications upon prediction completion.
How It Works
Foldify simplifies the prediction process by breaking it down into easy, intuitive steps:
- Select a Prediction Model: Choose from the available tools: AlphaFold3, AlphaFold2, ColabFold, OmegaFold, or ESMFold.
- Insert Input Data: Provide an amino acid sequence. For AlphaFold3, you can also upload a JSON configuration file.
- Configure Parameters: Adjust the settings based on your preferences.
- Submit the Job: Your computation request will be processed in a Kubernetes cluster.
- Monitor Progress: Track the status of your job in real-time on the Dashboard.
- Download or Visualize Results: Once the job is complete, you can download the results or explore the 3D structure and computation parameters directly in the web interface.
Find more in our documentation.
Optimizing AlphaFold3 in Kubernetes
Our platform is deployed on a Kubernetes cluster, where all computation jobs run. However, protein prediction is computationally demanding, and shared memory within the cluster can be a limiting factor. To optimize AlphaFold3 computations, we restructured the execution of the pipeline in Kubernetes. Previously, the entire computation was executed within a single Kubernetes pod. Now, the pipeline is distributed across multiple pods, with two primary pods handling the CPU and GPU computations separately. The GPU pod is only spawned after the long-running CPU phase is completed, preventing inefficient GPU reservation while CPU tasks are still running. Furthermore, the CPU-intensive stage is further divided, ensuring each subprocess runs in its dedicated pod. The granularity allows for better resource utilization as completed pods release their allocated resources, thereby enhancing overall performance.
Conclusion
Foldify simplifies protein structure prediction, making it accessible, efficient, and scalable for everyone. Whether you are an experienced researcher or a student exploring structural biology, our platform provides you with the essential tools for success.
We invite you to try protein structure prediction with Foldify. Your feedback is invaluable in helping us improve and expand our platform.